Hello,
I am trying to import an .asc file using read_raw_eyelink():
raw = read_raw_eyelink(fname)
However, I get the following error:
Loading /Users/dani/Desktop/test.asc
Pixel coordinate data detected.
Pass `scalings=dict(eyegaze=1e3)` when using plot method to make traces more legible.
Pupil-size area reported.
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/internals/construction.py:934, in _finalize_columns_and_data(content, columns, dtype)
933 try:
--> 934 columns = _validate_or_indexify_columns(contents, columns)
935 except AssertionError as err:
936 # GH#26429 do not raise user-facing AssertionError
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/internals/construction.py:981, in _validate_or_indexify_columns(content, columns)
979 if not is_mi_list and len(columns) != len(content): # pragma: no cover
980 # caller's responsibility to check for this...
--> 981 raise AssertionError(
982 f"{len(columns)} columns passed, passed data had "
983 f"{len(content)} columns"
984 )
985 if is_mi_list:
986 # check if nested list column, length of each sub-list should be equal
AssertionError: 10 columns passed, passed data had 9 columns
The above exception was the direct cause of the following exception:
ValueError Traceback (most recent call last)
Cell In[4], line 2
1 fname = "/Users/dani/Desktop/test.asc"
----> 2 raw = read_raw_eyelink(fname)
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/mne/io/eyelink/eyelink.py:350, in read_raw_eyelink(fname, preload, verbose, create_annotations, apply_offsets, find_overlaps, overlap_threshold, gap_description)
342 if extension not in ".asc":
343 raise ValueError(
344 "This reader can only read eyelink .asc files."
345 f" Got extension {extension} instead. consult eyelink"
346 " manual for converting eyelink data format (.edf)"
347 " files to .asc format."
348 )
--> 350 return RawEyelink(
351 fname,
352 preload=preload,
353 verbose=verbose,
354 create_annotations=create_annotations,
355 apply_offsets=apply_offsets,
356 find_overlaps=find_overlaps,
357 overlap_threshold=overlap_threshold,
358 gap_desc=gap_description,
359 )
File :12, in __init__(self, fname, preload, verbose, create_annotations, apply_offsets, find_overlaps, overlap_threshold, gap_desc)
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/mne/io/eyelink/eyelink.py:457, in RawEyelink.__init__(self, fname, preload, verbose, create_annotations, apply_offsets, find_overlaps, overlap_threshold, gap_desc)
455 sfreq = _get_sfreq(self._event_lines["SAMPLES"][0])
456 col_names, ch_names = self._infer_col_names()
--> 457 self._create_dataframes(
458 col_names, sfreq, find_overlaps=find_overlaps, threshold=overlap_threshold
459 )
460 info = self._create_info(ch_names, sfreq)
461 eye_ch_data = self.dataframes["samples"][ch_names]
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/mne/io/eyelink/eyelink.py:745, in RawEyelink._create_dataframes(self, col_names, sfreq, find_overlaps, threshold)
742 first_samp = self._event_lines["START"][0][0]
744 # dataframe for samples
--> 745 self.dataframes["samples"] = pd.DataFrame(
746 self._sample_lines, columns=col_names["sample"]
747 )
748 if "HREF" in self._rec_info:
749 pos_names = (
750 EYELINK_COLS["pos"]["left"][:-1] + EYELINK_COLS["pos"]["right"][:-1]
751 )
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/frame.py:782, in DataFrame.__init__(self, data, index, columns, dtype, copy)
780 if columns is not None:
781 columns = ensure_index(columns)
--> 782 arrays, columns, index = nested_data_to_arrays(
783 # error: Argument 3 to "nested_data_to_arrays" has incompatible
784 # type "Optional[Collection[Any]]"; expected "Optional[Index]"
785 data,
786 columns,
787 index, # type: ignore[arg-type]
788 dtype,
789 )
790 mgr = arrays_to_mgr(
791 arrays,
792 columns,
(...)
795 typ=manager,
796 )
797 else:
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/internals/construction.py:498, in nested_data_to_arrays(data, columns, index, dtype)
495 if is_named_tuple(data[0]) and columns is None:
496 columns = ensure_index(data[0]._fields)
--> 498 arrays, columns = to_arrays(data, columns, dtype=dtype)
499 columns = ensure_index(columns)
501 if index is None:
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/internals/construction.py:840, in to_arrays(data, columns, dtype)
837 data = [tuple(x) for x in data]
838 arr = _list_to_arrays(data)
--> 840 content, columns = _finalize_columns_and_data(arr, columns, dtype)
841 return content, columns
File ~/anaconda3/envs/mne-env/lib/python3.11/site-packages/pandas/core/internals/construction.py:937, in _finalize_columns_and_data(content, columns, dtype)
934 columns = _validate_or_indexify_columns(contents, columns)
935 except AssertionError as err:
936 # GH#26429 do not raise user-facing AssertionError
--> 937 raise ValueError(err) from err
939 if len(contents) and contents[0].dtype == np.object_:
940 contents = convert_object_array(contents, dtype=dtype)
ValueError: 10 columns passed, passed data had 9 columns
My operating system, MNE version, and Python version are:
- MNE version: 1.4.2
- operating system: macOS-13.4.1-arm64-arm-64bit
- Python: 3.11.4
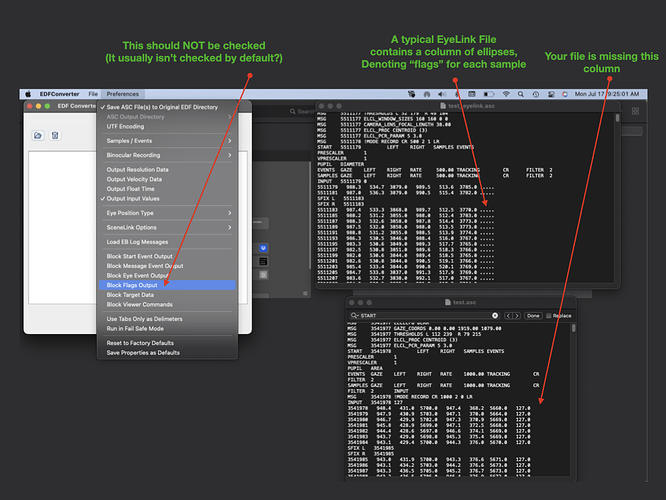
Has anyone ever stumbled upon something like this? The eye tracking file itself is not corrupted (I can import it in RStudio and it looks fine).
This is a link to a test file: https://drive.google.com/file/d/1QQbimGsPpc58WgJCQ91K9AyQ2oveNzpA/view?usp=sharing
Thanks so much and best wishes,
Dani