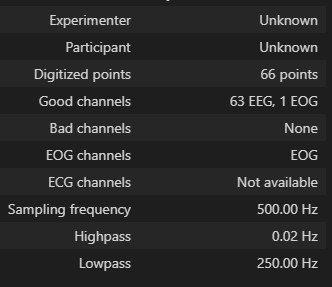
Hi.
I ran the code at the bottom and got an error code.
I’m totally new to eeg data analysis, so I really have no idea where the error came from.
I could only guess there is something wrong with data itself, like wrong data format or wrong data info.
Can anyone help out to fix it or just guess what the problem is?
I’d appreciate it so much.
auto_noisy_chs, auto_flat_chs, auto_scores = mne.preprocessing.find_bad_channels_maxwell(
raw, return_scores=True, verbose=True)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[36], line 1
----> 1 auto_noisy_chs, auto_flat_chs, auto_scores = mne.preprocessing.find_bad_channels_maxwell(
2 raw_prep, return_scores=True, verbose=True)
File :10, in find_bad_channels_maxwell(raw, limit, duration, min_count, return_scores, origin, int_order, ext_order, calibration, cross_talk, coord_frame, regularize, ignore_ref, bad_condition, head_pos, mag_scale, skip_by_annotation, h_freq, extended_proj, verbose)
File...\mne\preprocessing\maxwell.py:2237, in find_bad_channels_maxwell(***failed resolving arguments***)
2234 min_count = min(_ensure_int(min_count, 'min_count'), len(starts))
2235 logger.info('Scanning for bad channels in %d interval%s (%0.1f sec) ...'
2236 % (len(starts), _pl(starts), step / raw.info['sfreq']))
-> 2237 params = _prep_maxwell_filter(
2238 raw, skip_by_annotation=[], # already accounted for
2239 origin=origin, int_order=int_order, ext_order=ext_order,
2240 calibration=calibration, cross_talk=cross_talk,
2241 coord_frame=coord_frame, regularize=regularize,
2242 ignore_ref=ignore_ref, bad_condition=bad_condition, head_pos=head_pos,
2243 mag_scale=mag_scale, extended_proj=extended_proj)
2244 del origin, int_order, ext_order, calibration, cross_talk, coord_frame
2245 del regularize, ignore_ref, bad_condition, head_pos, mag_scale
File :12, in _prep_maxwell_filter(raw, origin, int_order, ext_order, calibration, cross_talk, st_duration, st_correlation, coord_frame, destination, regularize, ignore_ref, bad_condition, head_pos, st_fixed, st_only, mag_scale, skip_by_annotation, extended_proj, reconstruct, verbose)
File ...\mne\preprocessing\maxwell.py:412, in _prep_maxwell_filter(raw, origin, int_order, ext_order, calibration, cross_talk, st_duration, st_correlation, coord_frame, destination, regularize, ignore_ref, bad_condition, head_pos, st_fixed, st_only, mag_scale, skip_by_annotation, extended_proj, reconstruct, verbose)
...
--> 493 raise ValueError('No channels match the selection.')
494 n_unique = len(np.unique(np.arange(len(info['ch_names']))[sel]))
495 if n_unique != len(sel):
ValueError: No channels match the selection.