MNE version: 1.7.1
Python version : 3.12
Operating system: Win10
Hello everyone,
I’m currently performing a time-frequency analysis on EEG data with a within-subject design (19 participants, two conditions: pre-TMS and post-TMS).
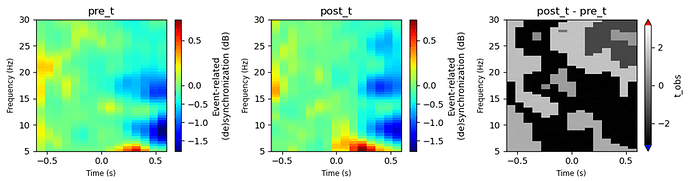
Here’s what we have done so far: we applied an STFT on epochs on [-1, 2]s around the events of interest for each subject. Then, we cropped the data between -0.6 and 1.5 s and applied a baseline on [-0.6, 0]s. We focus on a cluster of channels, so the resulting TFR was averaged on the epochs dimension and the channels included in this cluster, then normalized to dB.
At the group level, we want to compare the two conditions (pre vs. post) using the function mne.stats.spatio_temporal_cluster_1samp_test on the contrasted power between both conditions (TFR_post – TFR_pre), using a t-test as the core statistic. We kept classic permutation settings (n_permutations = 10000, step_down_p = 0.05) and used a TFCE threshold defined as threshold=dict(start=0, step=0.2). The adjacency has been defined as follows: mne.stats.combine_adjacency(len(times), len(freqs), 1)withspace_dim = 1 as we are working in the channel cluster).
My issue is that, when I visualize the contrasted TFR with the T-obs from the permutations test, I see large regions with equal t-values but no voxel is marked as significant. I’m wondering if the very low variability of the T-obs across the voxels is normal or not.
If I change the threshold (e.g., threshold=None or use a fixed numeric value), the T-obs all become 1 or -1.
We have already checked the shape of the TFR data array ([n_times, n_freqs]), and the contrast array ([n_subjects, n_times, n_freqs, n_channels], the one we use for the permutations test), and both look correct. The shape of the significant mask array we apply is also correct. We have cheked the data themselves and we are sure that we do not have any issue with the subject-level TFRs. It’s not a visualization problem either because we verified the settings of the configuration (lower/upper origin or other plt.imshow settings) of the plot.
We also reviewed the documentation, the forum (https://mne.discourse.group/t/cluster-based-permutation-tests-on-spatial-data/4572/2, https://mne.discourse.group/t/issue-with-tfce/2167, https://mne.discourse.group/t/issue-with-tfce/2167, https://mne.tools/1.7/auto_tutorials/stats-sensor-space/10_background_stats.html#tfce-example) and the relevant MNE tutorials but couldn’t find the source of the issue.
Finally, we are wondering if the T-obs output represents t-values from the first “layer” of permutation or the corrected sums of t-values after TFCE clustering? (the documentation is not entirely clear on this point, but it seems it should be the TFCE values).
Any insights or suggestions would be greatly appreciated, thank you for your help!
Ugo